Unraveling the Role of Anoikis in Non-Alcoholic Fatty Liver Disease Progression and Immune Cell Infiltration
DOI:
https://doi.org/10.71321/p63ws623Keywords:
NAFLD, Anoikis, Apoptosis, Machine learningAbstract
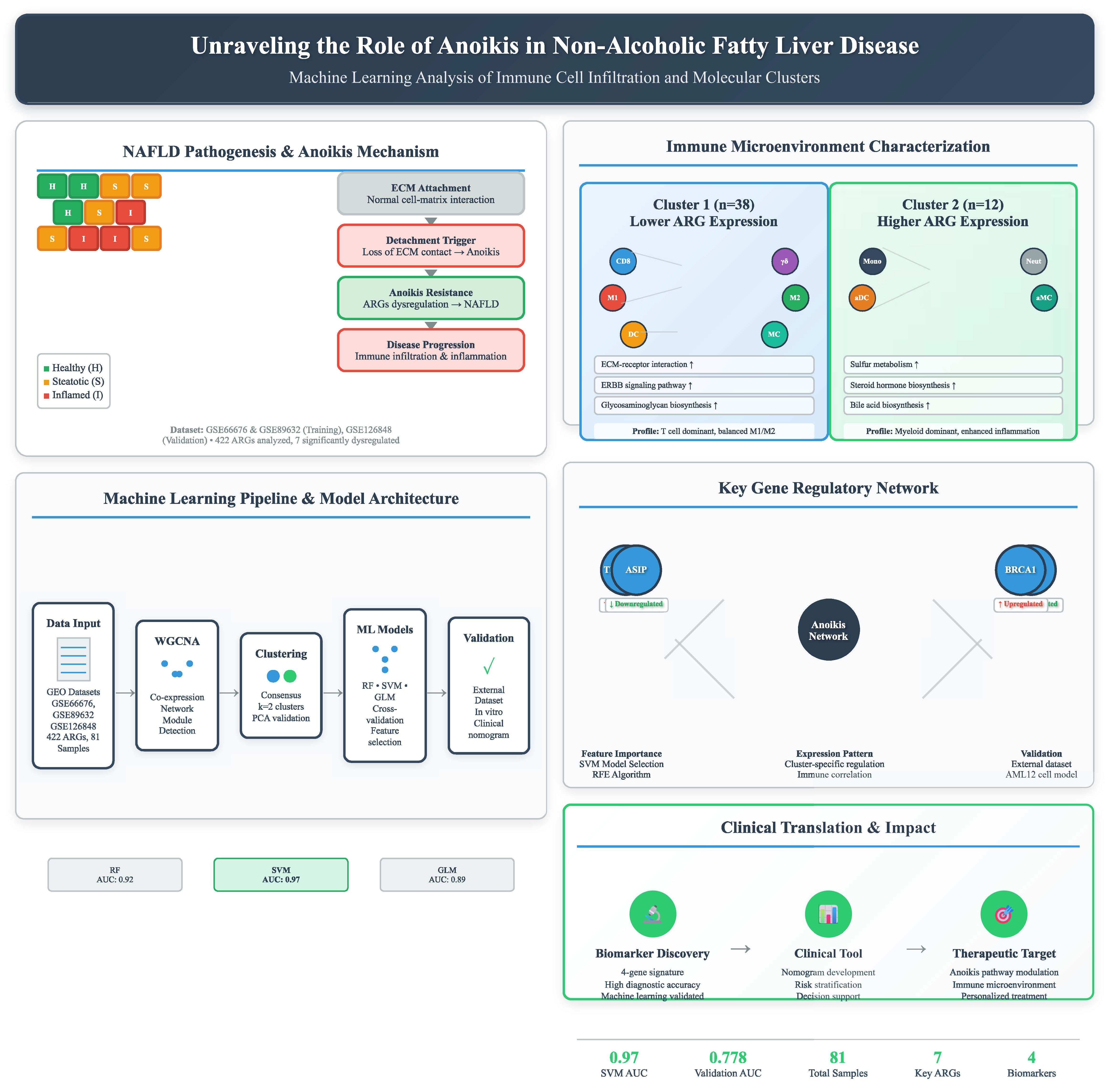
Background: Non-alcoholic fatty liver disease (NAFLD) is a prevalent chronic liver disease with complex molecular mechanisms. Anoikis, a distinct form of programmed cell death, has been implicated in disease progression, but its specific role in NAFLD remains unclear. This study aims to identify anoikis-related molecular clusters, explore their immune characteristics, and construct a predictive model for NAFLD prognosis.
Methods: Gene expression profiles of NAFLD samples were obtained from the Gene Expression Omnibus (GEO) database. Weighted gene co-expression network analysis (WGCNA) was applied to identify cluster-specific differentially expressed genes. Immune infiltration analysis was conducted to evaluate the association between anoikis-related clusters and immune cell composition. Machine learning was used to screen feature genes, and a predictive model was developed. The model’s performance was assessed using nomograms, calibration curves, and decision curve analysis (DCA).
Results: Two distinct anoikis-related molecular clusters were identified, each exhibiting unique immune microenvironment characteristics. Cluster 1 showed higher levels of CD8 T cells, γ-delta T cells, and macrophages (M1 and M2), while Cluster 2 had increased monocytes, activated dendritic cells, and neutrophils, reflecting inflammatory heterogeneity. Four key genes (TMEM169, THBS1, ASIP, and BRCA1) were identified through machine learning and incorporated into a predictive model. The model’s accuracy in predicting NAFLD prognosis was confirmed through nomograms, calibration curves, and DCA.
Conclusion: This study established an anoikis-related prognostic model for NAFLD and identified key genes involved in disease progression. The findings provide novel insights into the interplay between anoikis, immune responses, and NAFLD, offering potential biomarkers and therapeutic targets for personalized treatment.
References
[1] Guo X, Yin X, Liu Z, Wang J. Non-Alcoholic Fatty Liver Disease (NAFLD) Pathogenesis and Natural Products for Prevention and Treatment. Int J Mol Sci. 2022; https://doi.org/10.3390/ijms232415489
[2] Younossi Z. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention; https://doi.org/10.1038/nrgastro.2017.109
[3] Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatol Baltim Md. 2016;64:73–84; https://doi.org/10.1002/hep.28431
[4] Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol. 2015;62:S47-64; https://doi.org/10.1016/j.jhep.2014.12.012
[5] Li Y, Pan Q, Cheng M, Wu Z. Identification and validation of anoikis-associated gene SNCG as a prognostic biomarker in gastric cancer. Aging. 2023;15:2541–53; https://doi.org/10.1016/j.jhep.2014.12.012
[6] Ni K, Meng L. Mechanism of PANoptosis in metabolic dysfunction-associated steatotic liver disease. Clin Res Hepatol Gastroenterol. 2024;48:102381; DOI:10.1016/j.clinre.2024.102381
[7] Sun H-J, Jiao B, Wang Y, Zhang Y-H, Chen G, Wang Z-X, et al. Necroptosis contributes to non-alcoholic fatty liver disease pathoetiology with promising diagnostic and therapeutic functions. World J Gastroenterol. 2024;30:1968–81; https://doi.org/10.3748/wjg.v30.i14.1968
[8] Li R, Xue W, Wei H, Fan Q, Li X, Qiu Y, et al. Research Progress of Pyroptosis in Fatty Liver Disease. Int J Mol Sci. 2023;24:13065.
[9] Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559; https://doi.org/10.3390/ijms241713065
[10] Rigatti SJ. Random Forest. J Insur Med N Y N. 2017;47:31–9; https://doi.org/10.17849/insm-47-01-31-39.1
[11] Tan M, Pu J, Zheng B. Optimization of breast mass classification using sequential forward floating selection (SFFS) and a support vector machine (SVM) model. Int J Comput Assist Radiol Surg. 2014;9:1005–20; https://doi.org/10.1007/s11548-014-0992-1
[12] Weng G, Clark K, Akbarian A, Noudoost B, Nategh N. Time-varying generalized linear models: characterizing and decoding neuronal dynamics in higher visual areas. Front Comput Neurosci. 2024;18:1273053; https://doi.org/10.3389/fncom.2024.1273053
[13] Chu MJJ, Hickey AJR, Tagaloa S, Zhang L, Dare AJ, MacDonald JR, et al. Ob/ob mouse livers show decreased oxidative phosphorylation efficiencies and anaerobic capacities after cold ischemia. PloS One. 2014;9:e100609; https://doi.org/10.1371/journal.pone.0100609
[14] Roychowdhury S, McCullough RL, Sanz-Garcia C, Saikia P, Alkhouri N, Matloob A, et al. Receptor interacting protein 3 protects mice from high-fat diet-induced liver injury. Hepatol Baltim Md. 2016;64:1518–33; https://doi.org/10.1002/hep.28676
[15] Kazankov K, Jørgensen SMD, Thomsen KL, Møller HJ, Vilstrup H, George J, et al. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat Rev Gastroenterol Hepatol. 2019;16:145–59; https://doi.org/10.1038/s41575-018-0082-x
[16] Wang Y, Smith W, Hao D, He B, Kong L. M1 and M2 macrophage polarization and potentially therapeutic naturally occurring compounds. Int Immunopharmacol. 2019;70:459–66; https://doi.org/10.1016/j.intimp.2019.02.050
[17] Gong H, He Q, Zhu L, Feng Z, Sun M, Jiang J, et al. Associations between systemic inflammation indicators and nonalcoholic fatty liver disease: evidence from a prospective study. Front Immunol. 2024;15:1389967. https://doi.org/10.3389/fimmu.2024.1389967
[18] Wang Y-F, Zhang W-L, Li Z-X, Liu Y, Tan J, Yin H-Z, et al. METTL14 downregulation drives S100A4+ monocyte-derived macrophages via MyD88/NF-κB pathway to promote MAFLD progression. Signal Transduct Target Ther. 2024;9:91; https://doi.org/10.1038/s41392-024-01797-1
[19] Liu Z, Huang H, Ruan J, Wang Z, Xu C. The sulfur microbial diet and risk of nonalcoholic fatty liver disease: a prospective gene-diet study from the UK Biobank. Am J Clin Nutr. 2024;119:417–24; https://doi.org/10.1016/j.ajcnut.2023.11.012
[20] Bin D-H, Liu F, Peng K-P, Zhan M, Tan Y, Liu Q, et al. The relationship between follicle-stimulating hormone and metabolic dysfunction-associated fatty liver disease in men. Nutr Diabetes. 2024;14:52; https://doi.org/10.1038/s41387-024-00314-1
[21] Deo RC. Machine Learning in Medicine. Circulation. 2015;132:1920–30. https://doi.org/10.1161/CIRCULATIONAHA.115.001593
[22] Bai J, Xia M, Xue Y, Ma F, Cui A, Sun Y, et al. Thrombospondin 1 improves hepatic steatosis in diet-induced insulin-resistant mice and is associated with hepatic fat content in humans. EBioMedicine. 2020;57:102849; https://doi.org/10.1016/j.ebiom.2020.102849
[23] Mei J, Wang R, Xia D, Yang X, Zhou W, Wang H, et al. BRCA1 Is a Novel Prognostic Indicator and Associates with Immune Cell Infiltration in Hepatocellular Carcinoma. DNA Cell Biol. 2020;39:1838–49. https://doi.org/10.1089/dna.2020.5644
Type
Published
Data Availability Statement
The datasets presented in this study can be found in online repositories. Some data can be acquired in authors.
Issue
Section
License
Copyright (c) 2025 Life Conflux

This work is licensed under a Creative Commons Attribution 4.0 International License.







