Cell type- and developmental stage-specific mapping of polygenic risk across schizophrenia, depression, and bipolar disorder
DOI:
https://doi.org/10.71321/ftnk8985Keywords:
Psychiatric disorders, Genome-wide association study, Single-nucleus RNA sequencing, Cell type-specific risk, Neurodevelopmental trajectoriesAbstract
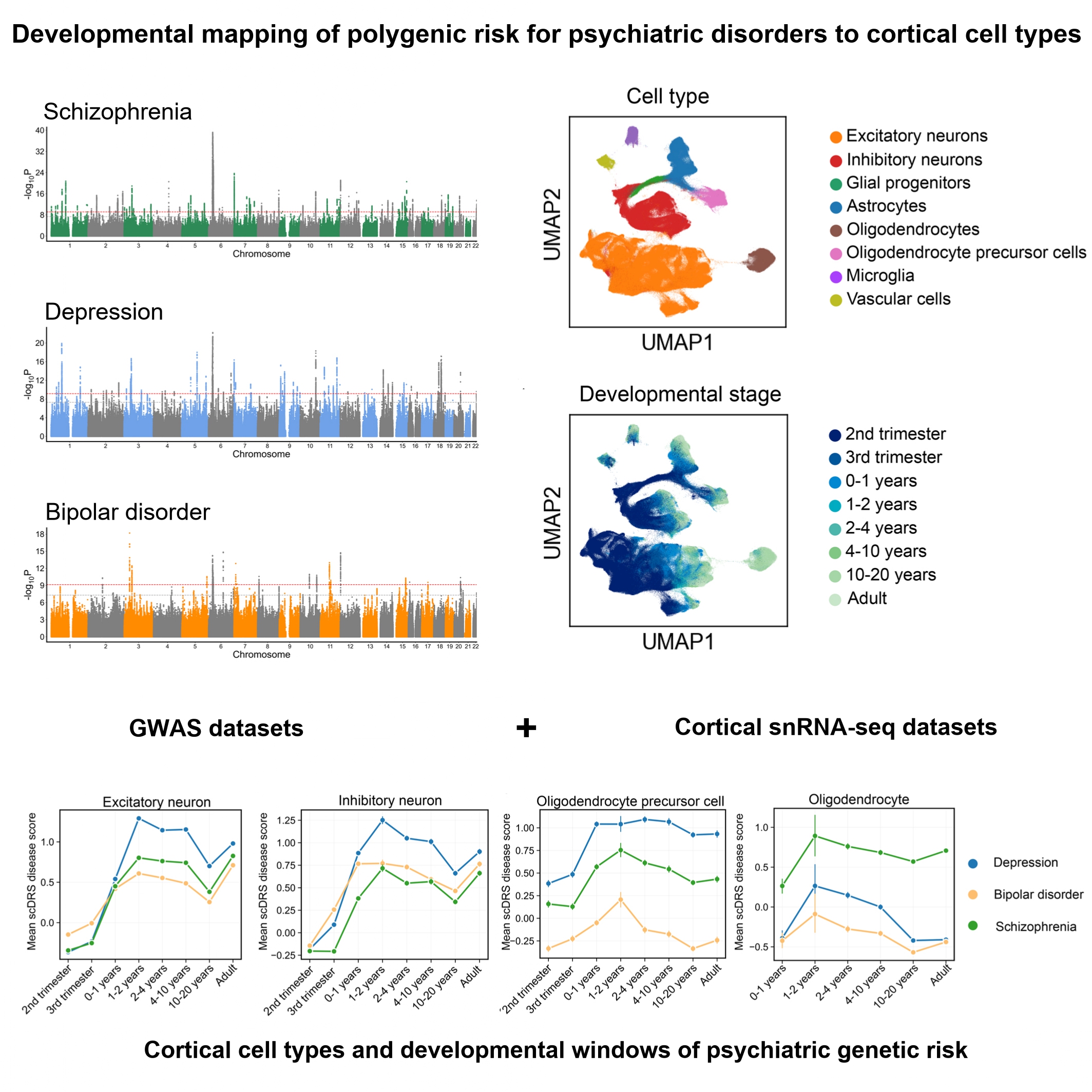
Schizophrenia, depression, and bipolar disorder are highly heritable psychiatric illnesses that share overlapping symptoms but also exhibit disorder-specific features. To dissect the cellular and developmental mechanisms of genetic risk, we integrated large-scale genome-wide association study (GWAS) data with human cortical single-nucleus RNA sequencing (snRNA-seq) data spanning gestation to adulthood (>700,000 nuclei from 106 donors). Gene-based analyses revealed 104 shared genes across disorders and convergent enrichment in synaptic pathways, alongside disorder-specific signals such as metal ion transport in schizophrenia. Using the single-cell disease relevance score (scDRS), we mapped polygenic risk across cortical cell types and developmental windows. Excitatory neurons were consistently implicated across all disorders from postnatal stages through adulthood, while inhibitory neurons showed broader vulnerability in depression and bipolar disorder, extending into the fetal period. Glial cells demonstrated disorder specificity: astrocytes were implicated across disorders during early postnatal synaptogenesis, oligodendrocyte precursor cells (OPCs) showed prolonged associations in depression, and mature oligodendrocytes were uniquely implicated in schizophrenia during childhood. These findings highlighted excitatory-inhibitory imbalance as a shared mechanism, alongside distinct glial and developmental trajectories contributing to disorder-specific pathophysiology. Our findings help to highlight the cortical cell types and developmental windows through which psychiatric genetic risk may act, offering insights into potential critical periods for intervention.
References
[1] Charlson F, van Ommeren M, Flaxman A, Cornett J, Whiteford H, & Saxena S. (2019). New WHO prevalence estimates of mental disorders in conflict settings: a systematic review and meta-analysis. Lancet, 394(10194), 240-248. https://doi.org/10.1016/s0140-6736(19)30934-1
[2] Li W, Yang Y, An FR, Zhang L, Ungvari GS, Jackson T, et al. (2020). Prevalence of comorbid depression in schizophrenia: A meta-analysis of observational studies. J Affect Disord, 273, 524-531. https://doi.org/10.1016/j.jad.2020.04.056
[3] Laursen TM, Agerbo E, & Pedersen CB. (2009). Bipolar disorder, schizoaffective disorder, and schizophrenia overlap: a new comorbidity index. J Clin Psychiatry, 70(10), 1432-1438. https://doi.org/10.4088/JCP.08m04807
[4] Waddington JL. (2024). From operational diagnostic to dimensional-continuum concepts of psychotic and non-psychotic illness: Embracing catatonia across psychopathology and intrinsic movement disorder in neural network dysfunction. Schizophr Res, 263, 99-108. https://doi.org/10.1016/j.schres.2022.10.001
[5] Schottner Sieler M, Golay P, Vieira S, Alameda L, Conus P, Klauser P, et al. (2025). A dimensional approach to psychosis: identifying cognition, depression, and thought disorder factors in a clinical sample. Schizophrenia (Heidelb), 11(1), 97. https://doi.org/10.1038/s41537-025-00641-x
[6] Li J, Long Z, Ji GJ, Han S, Chen Y, Yao G, et al. (2025). Major depressive disorder on a neuromorphic continuum. Nat Commun, 16(1), 2405. https://doi.org/10.1038/s41467-025-57682-0
[7] Trubetskoy V, Pardinas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, et al. (2022). Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature, 604(7906), 502-508. https://doi.org/10.1038/s41586-022-04434-5
[8] Als TD, Kurki MI, Grove J, Voloudakis G, Therrien K, Tasanko E, et al. (2023). Depression pathophysiology, risk prediction of recurrence and comorbid psychiatric disorders using genome-wide analyses. Nat Med, 29(7), 1832-1844. https://doi.org/10.1038/s41591-023-02352-1
[9] Mullins N, Forstner AJ, O'Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. (2021). Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet, 53(6), 817-829. https://doi.org/10.1038/s41588-021-00857-4
[10] Cross-Disorder Group of the Psychiatric Genomics Consortium. Electronic address pmhe, & Cross-Disorder Group of the Psychiatric Genomics C. (2019). Genomic Relationships, Novel Loci, and Pleiotropic Mechanisms across Eight Psychiatric Disorders. Cell, 179(7), 1469-1482 e1411. https://doi.org/10.1016/j.cell.2019.11.020
[11] Lee PH, Feng YA, & Smoller JW. (2021). Pleiotropy and Cross-Disorder Genetics Among Psychiatric Disorders. Biol Psychiatry, 89(1), 20-31. https://doi.org/10.1016/j.biopsych.2020.09.026
[12] Cross-Disorder Group of the Psychiatric Genomics C. (2013). Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet, 381(9875), 1371-1379. https://doi.org/10.1016/S0140-6736(12)62129-1
[13] Sullivan PF, & Geschwind DH. (2019). Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell, 177(1), 162-183. https://doi.org/10.1016/j.cell.2019.01.015
[14] van Erp TG, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. (2016). Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry, 21(4), 547-553. https://doi.org/10.1038/mp.2015.63
[15] Schmaal L, Pozzi E, T CH, van Velzen LS, Veer IM, Opel N, et al. (2020). ENIGMA MDD: seven years of global neuroimaging studies of major depression through worldwide data sharing. Transl Psychiatry, 10(1), 172. https://doi.org/10.1038/s41398-020-0842-6
[16] Hibar DP, Westlye LT, Doan NT, Jahanshad N, Cheung JW, Ching CRK, et al. (2018). Cortical abnormalities in bipolar disorder: an MRI analysis of 6503 individuals from the ENIGMA Bipolar Disorder Working Group. Mol Psychiatry, 23(4), 932-942. https://doi.org/10.1038/mp.2017.73
[17] Rootes-Murdy K, Panta S, Kelly R, Romero J, Quidé Y, Cairns MJ, et al. (2024). Cortical similarities in psychiatric and mood disorders identified in federated VBM analysis via COINSTAC. Patterns (N Y), 5(7), 100987. https://doi.org/10.1016/j.patter.2024.100987
[18] Birnbaum R, & Weinberger DR. (2017). Genetic insights into the neurodevelopmental origins of schizophrenia. Nat Rev Neurosci, 18(12), 727-740. https://doi.org/10.1038/nrn.2017.125
[19] Cameron D, Mi D, Vinh NN, Webber C, Li M, Marin O, et al. (2023). Single-Nuclei RNA Sequencing of 5 Regions of the Human Prenatal Brain Implicates Developing Neuron Populations in Genetic Risk for Schizophrenia. Biol Psychiatry, 93(2), 157-166. https://doi.org/10.1016/j.biopsych.2022.06.033
[20] Li M, Santpere G, Imamura Kawasawa Y, Evgrafov OV, Gulden FO, Pochareddy S, et al. (2018). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science, 362(6420). https://doi.org/10.1126/science.aat7615
[21] Schork AJ, Won H, Appadurai V, Nudel R, Gandal M, Delaneau O, et al. (2019). A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat Neurosci, 22(3), 353-361. https://doi.org/10.1038/s41593-018-0320-0
[22] Herring CA, Simmons RK, Freytag S, Poppe D, Moffet JJD, Pflueger J, et al. (2022). Human prefrontal cortex gene regulatory dynamics from gestation to adulthood at single-cell resolution. Cell, 185(23), 4428-4447 e4428. https://doi.org/10.1016/j.cell.2022.09.039
[23] Velmeshev D, Perez Y, Yan Z, Valencia JE, Castaneda-Castellanos DR, Wang L, et al. (2023). Single-cell analysis of prenatal and postnatal human cortical development. Science, 382(6667), eadf0834. https://doi.org/10.1126/science.adf0834
[24] Bryois J, Skene NG, Hansen TF, Kogelman LJA, Watson HJ, Liu Z, et al. (2020). Genetic identification of cell types underlying brain complex traits yields insights into the etiology of Parkinson's disease. Nat Genet, 52(5), 482-493. https://doi.org/10.1038/s41588-020-0610-9
[25] Jagadeesh KA, Dey KK, Montoro DT, Mohan R, Gazal S, Engreitz JM, et al. (2022). Identifying disease-critical cell types and cellular processes by integrating single-cell RNA-sequencing and human genetics. Nat Genet, 54(10), 1479-1492. https://doi.org/10.1038/s41588-022-01187-9
[26] Zhang MJ, Hou K, Dey KK, Sakaue S, Jagadeesh KA, Weinand K, et al. (2022). Polygenic enrichment distinguishes disease associations of individual cells in single-cell RNA-seq data. Nat Genet, 54(10), 1572-1580. https://doi.org/10.1038/s41588-022-01167-z
[27] Wolf FA, Angerer P, & Theis FJ. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol, 19(1), 15. https://doi.org/10.1186/s13059-017-1382-0
[28] de Leeuw CA, Mooij JM, Heskes T, & Posthuma D. (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol, 11(4), e1004219. https://doi.org/10.1371/journal.pcbi.1004219
[29] Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. (2015). A global reference for human genetic variation. Nature, 526(7571), 68-74. https://doi.org/10.1038/nature15393
[30] Chen J, Bardes EE, Aronow BJ, & Jegga AG. (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res, 37(Web Server issue), W305-311. https://doi.org/10.1093/nar/gkp427
[31] Healy J, & McInnes L. (2024). Uniform manifold approximation and projection. Nature Reviews Methods Primers, 4(1), 82. https://doi.org/10.1038/s43586-024-00363-x
[32] Petanjek Z, Judas M, Simic G, Rasin MR, Uylings HB, Rakic P, et al. (2011). Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc Natl Acad Sci U S A, 108(32), 13281-13286. https://doi.org/10.1073/pnas.1105108108
[33] Huttenlocher PR, & Dabholkar AS. (1997). Regional differences in synaptogenesis in human cerebral cortex. J Comp Neurol, 387(2), 167-178. https://doi.org/10.1002/(sici)1096-9861(19971020)387:2<167::aid-cne1>3.0.co;2-z
[34] Clarke LE, & Barres BA. (2013). Emerging roles of astrocytes in neural circuit development. Nat Rev Neurosci, 14(5), 311-321. https://doi.org/10.1038/nrn3484
[35] Molofsky AV, & Deneen B. (2015). Astrocyte development: A Guide for the Perplexed. Glia, 63(8), 1320-1329. https://doi.org/10.1002/glia.22836
[36] Zhou B, Zhu Z, Ransom BR, & Tong X. (2021). Oligodendrocyte lineage cells and depression. Mol Psychiatry, 26(1), 103-117. https://doi.org/10.1038/s41380-020-00930-0
[37] Douaud G, Smith S, Jenkinson M, Behrens T, Johansen-Berg H, Vickers J, et al. (2007). Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain, 130(Pt 9), 2375-2386. https://doi.org/10.1093/brain/awm184
[38] Takahashi N, Sakurai T, Davis KL, & Buxbaum JD. (2011). Linking oligodendrocyte and myelin dysfunction to neurocircuitry abnormalities in schizophrenia. Prog Neurobiol, 93(1), 13-24. https://doi.org/10.1016/j.pneurobio.2010.09.004
[39] Lin YC, & Koleske AJ. (2010). Mechanisms of synapse and dendrite maintenance and their disruption in psychiatric and neurodegenerative disorders. Annu Rev Neurosci, 33, 349-378. https://doi.org/10.1146/annurev-neuro-060909-153204
[40] Howes OD, & Shatalina E. (2022). Integrating the Neurodevelopmental and Dopamine Hypotheses of Schizophrenia and the Role of Cortical Excitation-Inhibition Balance. Biol Psychiatry, 92(6), 501-513. https://doi.org/10.1016/j.biopsych.2022.06.017
[41] Hu YT, Tan ZL, Hirjak D, & Northoff G. (2023). Brain-wide changes in excitation-inhibition balance of major depressive disorder: a systematic review of topographic patterns of GABA- and glutamatergic alterations. Mol Psychiatry, 28(8), 3257-3266. https://doi.org/10.1038/s41380-023-02193-x
[42] Lee Y, Zhang Y, Kim S, & Han K. (2018). Excitatory and inhibitory synaptic dysfunction in mania: an emerging hypothesis from animal model studies. Exp Mol Med, 50(4), 1-11. https://doi.org/10.1038/s12276-018-0028-y
[43] Kelly S, Jahanshad N, Zalesky A, Kochunov P, Agartz I, Alloza C, et al. (2018). Widespread white matter microstructural differences in schizophrenia across 4322 individuals: results from the ENIGMA Schizophrenia DTI Working Group. Mol Psychiatry, 23(5), 1261-1269. https://doi.org/10.1038/mp.2017.170
Type
Published
Data Availability Statement
The GWAS summary statistics for schizophrenia, depression, and bipolar disorder are available from the Psychiatric Genomics Consortium (https://pgc.unc.edu/). The single-nucleus RNA sequencing dataset of prenatal and postnatal human cortical development is available from the original publication (Velmeshev et al., Science, 2019).
Issue
Section
License
Copyright (c) 2025 Brain Conflux

This work is licensed under a Creative Commons Attribution 4.0 International License.







