Research Progress of Epigenetic Regulation in Stroke Treatment
DOI:
https://doi.org/10.71321/y2sx8j49Keywords:
Epigenetic regulation; , Stroke;, DNA methylation;, Histone modification;, Non-coding RNA;, Therapeutic targetsAbstract
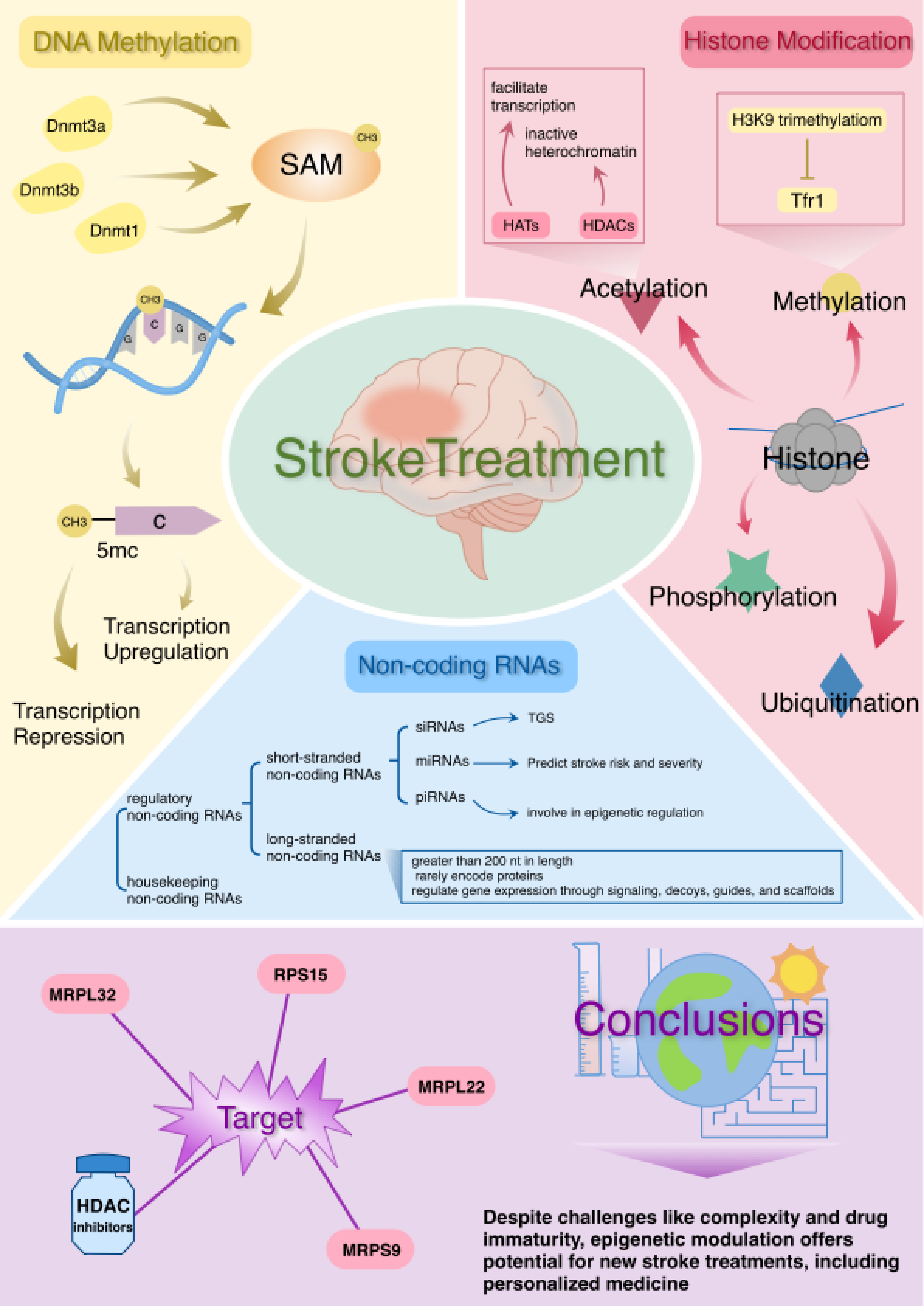
Cerebrovascular diseases are a serious threat to human health, among which stroke is extremely harmful. In recent years, more and more studies have shown that epigenetic regulation is crucial in the development of cerebrovascular diseases. In this review, we focus on the research progress of epigenetic regulation in stroke treatment, detailing the regulatory mechanisms of DNA methylation, histone modification and non-coding RNAs, and analyzing their roles in the pathophysiology of stroke. It was found that ischemic stroke causes dynamic changes in DNA methylation, which affects gene expression and alters the process of injury and recovery; histone modification levels are also altered after stroke, which affects chromatin status and gene transcription; and noncoding RNAs, such as miRNAs, siRNAs, piRNAs, and lncRNAs, which play a key role in gene expression regulation, are associated with the risk of stroke, severity and symptom onset. In addition, therapeutic strategies targeting epigenetic regulation are also discussed. Although facing challenges such as complex mechanisms, susceptibility to environmental influences, and the early stage of drug development, epigenetic regulation is still very promising in the treatment of stroke, and it is expected to provide a new theoretical basis and research direction for the prevention and treatment of stroke in the future.
References
[1] Abe A, Tanaka M, Yasuoka A, Saito Y, Okada S, Mishina M, et al. (2020) Changes in Whole-Blood microRNA Profiles during the Onset and Treatment Process of Cerebral Infarction: A Human Study. Int J Mol Sci, 21(9). https://doi.org/10.3390/ijms21093107
[2] Amaral PP, Dinger ME, Mercer TR, Mattick JS. (2008) The eukaryotic genome as an RNA machine. Science, 319(5871)1787-9. https://doi.org/10.1126/science.1155472
[3] Baccarelli A, Tarantini L, Wright RO, Bollati V, Litonjua AA, Zanobetti A, et al. (2010) Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA normative aging study. Epigenetics, 5(3)222-8. https://doi.org/10.4161/epi.5.3.11377
[4] Bakhit Y, Schmitt I, Hamed A, Ibrahim EAA, Mohamed IN, El-Sadig SM, et al. (2022) Methylation of alpha-synuclein in a Sudanese cohort. Parkinsonism Relat Disord, 1016-8. https://doi.org/10.1016/j.parkreldis.2022.05.009
[5] Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. (2007) High-resolution profiling of histone methylations in the human genome. Cell, 129(4)823-37. https://doi.org/10.1016/j.cell.2007.05.009
[6] Basavarajappa BS, Subbanna S. (2021) Histone Methylation Regulation in Neurodegenerative Disorders. Int J Mol Sci, 22(9). https://doi.org/10.3390/ijms22094654
[7] Bayne EH, Allshire RC. (2005) RNA-directed transcriptional gene silencing in mammals. Trends Genet, 21(7)370-3. https://doi.org/10.1016/j.tig.2005.05.007
[8] Cao X, Yeo G, Muotri AR, Kuwabara T, Gage FH. (2006) Noncoding RNAs in the mammalian central nervous system. Annu Rev Neurosci, 2977-103. https://doi.org/10.1146/annurev.neuro.29.051605.112839
[9] Carthew RW, Sontheimer EJ. (2009) Origins and Mechanisms of miRNAs and siRNAs. Cell, 136(4)642-55. https://doi.org/10.1016/j.cell.2009.01.035
[10] Chen J, Liu P, Dong X, Jin J, Xu Y. (2021) The role of lncRNAs in ischemic stroke. Neurochem Int, 147105019. https://doi.org/10.1016/j.neuint.2021.105019
[11] Collaborators GBDSRF. (2024) Global, regional, and national burden of stroke and its risk factors, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet Neurol, 23(10)973-1003. https://doi.org/10.1016/S1474-4422(24)00369-7
[12] Costa FF. (2008) Non-coding RNAs, epigenetics and complexity. Gene, 410(1)9-17. https://doi.org/10.1016/j.gene.2007.12.008
[13] Cullell N, Soriano-Tarraga C, Gallego-Fabrega C, Carcel-Marquez J, Torres-Aguila NP, Muino E, et al. (2022) DNA Methylation and Ischemic Stroke Risk: An Epigenome-Wide Association Study. Thromb Haemost, 122(10)1767-78. https://doi.org/10.1055/s-0042-1749328
[14] Demyanenko S, Berezhnaya E, Neginskaya M, Rodkin S, Dzreyan V, Pitinova M. (2019) capital ES, Cyrilliclass II histone deacetylases in the post-stroke recovery period-expression, cellular, and subcellular localization-promising targets for neuroprotection. J Cell Biochem, 120(12)19590-609. https://doi.org/10.1002/jcb.29266
[15] Demyanenko S, Neginskaya M, Berezhnaya E. (2018) Expression of Class I Histone Deacetylases in Ipsilateral and Contralateral Hemispheres after the Focal Photothrombotic Infarction in the Mouse Brain. Transl Stroke Res, 9(5)471-83. https://doi.org/10.1007/s12975-017-0595-6
[16] Demyanenko S, Uzdensky A. (2019) Epigenetic Alterations Induced by Photothrombotic Stroke in the Rat Cerebral Cortex: Deacetylation of Histone h3, Upregulation of Histone Deacetylases and Histone Acetyltransferases. Int J Mol Sci, 20(12). https://doi.org/10.3390/ijms20122882
[17] Deng P, Chen QM, Hong C, Wang CY. (2015) Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int J Oral Sci, 7(4)197-204. https://doi.org/10.1038/ijos.2015.41
[18] Feng J, Chang H, Li E, Fan G. (2005) Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res, 79(6)734-46. https://doi.org/10.1002/jnr.20404
[19] Fujii R, Yamada H, Munetsuna E, Yamazaki M, Mizuno G, Tsuboi Y, et al. (2019) Dietary vegetable intake is inversely associated with ATP-binding cassette protein A1 (ABCA1) DNA methylation levels among Japanese women. Nutrition, 651-5. https://doi.org/10.1016/j.nut.2019.02.010
[20] Gallego-Fabrega C, Carrera C, Reny JL, Fontana P, Slowik A, Pera J, et al. (2016) TRAF3 Epigenetic Regulation Is Associated With Vascular Recurrence in Patients With Ischemic Stroke. Stroke, 47(5)1180-6. https://doi.org/10.1161/STROKEAHA.115.012237
[21] Gediya P, Parikh PK, Vyas VK, Ghate MD. (2021) Histone deacetylase 2: A potential therapeutic target for cancer and neurodegenerative disorders. Eur J Med Chem, 216113332. https://doi.org/10.1016/j.ejmech.2021.113332
[22] Ghafouri-Fard S, Shoorei H, Taheri M. (2020) Non-coding RNAs participate in the ischemia-reperfusion injury. Biomed Pharmacother, 129110419. https://doi.org/10.1016/j.biopha.2020.110419
[23] Ghildiyal M, Zamore PD. (2009) Small silencing RNAs: an expanding universe. Nat Rev Genet, 10(2)94-108. https://doi.org/10.1038/nrg2504
[24] Goto K, Numata M, Komura JI, Ono T, Bestor TH, Kondo H. (1994) Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation, 56(1-2)39-44. https://doi.org/10.1046/j.1432-0436.1994.56120039.x
[25] Greer EL, Shi Y. (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet, 13(5)343-57. https://doi.org/10.1038/nrg3173
[26] Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res, 34(Database issue)D140-4. https://doi.org/10.1093/nar/gkj112
[27] Gurley LR, Walters RA, Tobey RA. (1973) Histone phosphorylation in late interphase and mitosis. Biochem Biophys Res Commun, 50(3)744-50. https://doi.org/10.1016/0006-291x(73)91307-7
[28] He S, Ye X, Duan R, Zhao Y, Wei Y, Wang Y, et al. (2022) Epigenome-Wide Association Study Reveals Differential Methylation Sites and Association of Gene Expression Regulation with Ischemic Moyamoya Disease in Adults. Oxid Med Cell Longev, 20227192060. https://doi.org/10.1155/2022/7192060
[29] Holliday R. (2006) Epigenetics: a historical overview. Epigenetics, 1(2)76-80. https://doi.org/10.4161/epi.1.2.2762
[30] Hu Z, Zhong B, Tan J, Chen C, Lei Q, Zeng L. (2017) The Emerging Role of Epigenetics in Cerebral Ischemia. Mol Neurobiol, 54(3)1887-905. https://doi.org/10.1007/s12035-016-9788-3
[31] Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. (2010) Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature, 466(7310)1129-33. https://doi.org/10.1038/nature09303
[32] Jhelum P, Karisetty BC, Kumar A, Chakravarty S. (2017) Implications of Epigenetic Mechanisms and their Targets in Cerebral Ischemia Models. Curr Neuropharmacol, 15(6)815-30. https://doi.org/10.2174/1570159X14666161213143907
[33] Jiang D, Wang Y, Shen Y, Xu Y, Zhu H, Wang J, et al. (2016) Estrogen and promoter methylation in the regulation of PLA2G7 transcription. Gene, 591(1)262-7. https://doi.org/10.1016/j.gene.2016.07.048
[34] Jinek M, Doudna JA. (2009) A three-dimensional view of the molecular machinery of RNA interference. Nature, 457(7228)405-12. https://doi.org/10.1038/nature07755
[35] Joensuu EI, Nieminen TT, Lotsari JE, Pavicic W, Abdel-Rahman WM, Peltomaki P. (2015) Methyltransferase expression and tumor suppressor gene methylation in sporadic and familial colorectal cancer. Genes Chromosomes Cancer, 54(12)776-87. https://doi.org/10.1002/gcc.22289
[36] Jones PA. (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet, 13(7)484-92. https://doi.org/10.1038/nrg3230
[37] Jones PA, Baylin SB. (2007) The epigenomics of cancer. Cell, 128(4)683-92. https://doi.org/10.1016/j.cell.2007.01.029
[38] Kassis H, Shehadah A, Chopp M, Zhang ZG. (2017) Epigenetics in Stroke Recovery. Genes (Basel), 8(3). https://doi.org/10.3390/genes8030089
[39] Kawasaki H, Taira K. (2004) Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature, 431(7005)211-7. https://doi.org/10.1038/nature02889
[40] Kim M, Long TI, Arakawa K, Wang R, Yu MC, Laird PW. (2010) DNA methylation as a biomarker for cardiovascular disease risk. PLoS One, 5(3)e9692. https://doi.org/10.1371/journal.pone.0009692
[41] Kozomara A, Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res, 42(Database issue)D68-73. https://doi.org/10.1093/nar/gkt1181
[42] Lan T, Hu L, Sun T, Wang X, Xiao Z, Shen D, et al. (2023) H3K9 trimethylation dictates neuronal ferroptosis through repressing Tfr1. J Cereb Blood Flow Metab, 43(8)1365-81. https://doi.org/10.1177/0271678X231165653
[43] Lanzillotta A, Pignataro G, Branca C, Cuomo O, Sarnico I, Benarese M, et al. (2013) Targeted acetylation of NF-kappaB/RelA and histones by epigenetic drugs reduces post-ischemic brain injury in mice with an extended therapeutic window. Neurobiol Dis, 49177-89. https://doi.org/10.1016/j.nbd.2012.08.018
[44] Li Y. (2021) Modern epigenetics methods in biological research. Methods, 187104-13. https://doi.org/10.1016/j.ymeth.2020.06.022
[45] Lin H. (2007) piRNAs in the germ line. Science, 316(5823)397. https://doi.org/10.1126/science.1137543
[46] Lu M, Dong X, Zhang Z, Li W, Khoshnam SE. (2020) Non-coding RNAs in Ischemic Stroke: Roles in the Neuroinflammation and Cell Death. Neurotox Res, 38(3)564-78. https://doi.org/10.1007/s12640-020-00236-7
[47] Luo M, Hao L, Hu F, Dong Y, Gou L, Zhang W, et al. (2015) MicroRNA profiles and potential regulatory pattern during the early stage of spermatogenesis in mice. Sci China Life Sci, 58(5)442-50. https://doi.org/10.1007/s11427-014-4737-8
[48] Maejima H, Okamura M, Inoue T, Takamatsu Y, Nishio T, Liu Y. (2023) Epigenetic modifications in the motor cortex caused by exercise or pharmacological inhibition of histone deacetylases (HDACs) after intracerebral hemorrhage (ICH). Brain Res, 1806148286. https://doi.org/10.1016/j.brainres.2023.148286
[49] Maes T, Mascaro C, Ortega A, Lunardi S, Ciceri F, Somervaille TC, et al. (2015) KDM1 histone lysine demethylases as targets for treatments of oncological and neurodegenerative disease. Epigenomics, 7(4)609-26. https://doi.org/10.2217/epi.15.9
[50] Mandumpala JJ, Baby S, Tom AA, Godugu C, Shankaraiah N. (2022) Role of histone demethylases and histone methyltransferases in triple-negative breast cancer: Epigenetic mnemonics. Life Sci, 292120321. https://doi.org/10.1016/j.lfs.2022.120321
[51] Mercer TR, Dinger ME, Mattick JS. (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet, 10(3)155-9. https://doi.org/10.1038/nrg2521
[52] Moazed D. (2009) Small RNAs in transcriptional gene silencing and genome defence. Nature, 457(7228)413-20. https://doi.org/10.1038/nature07756
[53] Morris KV, Chan SW, Jacobsen SE, Looney DJ. (2004) Small interfering RNA-induced transcriptional gene silencing in human cells. Science, 305(5688)1289-92. https://doi.org/10.1126/science.1101372
[54] Ng GY, Lim YA, Sobey CG, Dheen T, Fann DY, Arumugam TV. (2018) Epigenetic regulation of inflammation in stroke. Ther Adv Neurol Disord, 111756286418771815. https://doi.org/10.1177/1756286418771815
[55] Ponting CP, Oliver PL, Reik W. (2009) Evolution and functions of long noncoding RNAs. Cell, 136(4)629-41. https://doi.org/10.1016/j.cell.2009.02.006
[56] Rechtsteiner A, Ercan S, Takasaki T, Phippen TM, Egelhofer TA, Wang W, et al. (2010) The histone H3K36 methyltransferase MES-4 acts epigenetically to transmit the memory of germline gene expression to progeny. PLoS Genet, 6(9)e1001091. https://doi.org/10.1371/journal.pgen.1001091
[57] Ruvkun G. (2001) Molecular biology. Glimpses of a tiny RNA world. Science, 294(5543)797-9. https://doi.org/10.1126/science.1066315
[58] Schweizer S, Meisel A, Marschenz S. (2013) Epigenetic mechanisms in cerebral ischemia. J Cereb Blood Flow Metab, 33(9)1335-46. https://doi.org/10.1038/jcbfm.2013.93
[59] Sharma AR, Shashikiran U, Uk AR, Shetty R, Satyamoorthy K, Rai PS. (2020) Aberrant DNA methylation and miRNAs in coronary artery diseases and stroke: a systematic review. Brief Funct Genomics, 19(4)259-85. https://doi.org/10.1093/bfgp/elz043
[60] Stanzione R, Cotugno M, Bianchi F, Marchitti S, Forte M, Volpe M, et al. (2020) Pathogenesis of Ischemic Stroke: Role of Epigenetic Mechanisms. Genes (Basel), 11(1). https://doi.org/10.3390/genes11010089
[61] Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev, 12(5)599-606. https://doi.org/10.1101/gad.12.5.599
[62] Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, et al. (2009) Expression profile of MicroRNAs in young stroke patients. PLoS One, 4(11)e7689. https://doi.org/10.1371/journal.pone.0007689
[63] Tchurikov NA. (2005) Molecular mechanisms of epigenetics. Biochemistry (Mosc), 70(4)406-23. https://doi.org/10.1007/s10541-005-0131-2
[64] Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. (2006) The Polycomb group protein EZH2 directly controls DNA methylation. Nature, 439(7078)871-4. https://doi.org/10.1038/nature04431
[65] Wang KC, Chang HY. (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell, 43(6)904-14. https://doi.org/10.1016/j.molcel.2011.08.018
[66] Wang SW, Liu Z, Shi ZS. (2018) Non-Coding RNA in Acute Ischemic Stroke: Mechanisms, Biomarkers and Therapeutic Targets. Cell Transplant, 27(12)1763-77. https://doi.org/10.1177/0963689718806818
[67] Wang Y, Zhang X, Liu H, Zhou X. (2021) Chemical methods and advanced sequencing technologies for deciphering mRNA modifications. Chem Soc Rev, 50(24)13481-97. https://doi.org/10.1039/d1cs00920f
[68] Wang Z, Tsai LK, Munasinghe J, Leng Y, Fessler EB, Chibane F, et al. (2012) Chronic valproate treatment enhances postischemic angiogenesis and promotes functional recovery in a rat model of ischemic stroke. Stroke, 43(9)2430-6. https://doi.org/10.1161/STROKEAHA.112.652545
[69] Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. (2009) Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell, 138(5)1019-31. https://doi.org/10.1016/j.cell.2009.06.049
[70] Weake VM, Workman JL. (2008) Histone ubiquitination: triggering gene activity. Mol Cell, 29(6)653-63. https://doi.org/10.1016/j.molcel.2008.02.014
[71] Wei JW, Huang K, Yang C, Kang CS. (2017) Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep, 37(1)3-9. https://doi.org/10.3892/or.2016.5236
[72] Wolf PA, Grotta JC. (2000) Cerebrovascular disease. Circulation, 102(20 Suppl 4)IV75-80. https://doi.org/10.1161/01.cir.102.suppl_4.iv-75
[73] Yu H. (2009) [Epigenetics: advances of non-coding RNAs regulation in mammalian cells]. Yi Chuan, 31(11)1077-86. https://doi.org/10.3724/sp.j.1005.2009.01077
[74] Zaratiegui M, Irvine DV, Martienssen RA. (2007) Noncoding RNAs and gene silencing. Cell, 128(4)763-76. https://doi.org/10.1016/j.cell.2007.02.016
[75] Zhan L, Chen M, Pang T, Li X, Long L, Liang D, et al. (2023) Attenuation of Piwil2 induced by hypoxic postconditioning prevents cerebral ischemic injury by inhibiting CREB2 promoter methylation. Brain Pathol, 33(1)e13109. https://doi.org/10.1111/bpa.13109
[76] Zhao LY, Song J, Liu Y, Song CX, Yi C. (2020) Mapping the epigenetic modifications of DNA and RNA. Protein Cell, 11(11)792-808. https://doi.org/10.1007/s13238-020-00733-7
[77] Zhou W, Wang J, Man WY, Zhang QW, Xu WG. (2015) siRNA silencing EZH2 reverses cisplatin-resistance of human non-small cell lung and gastric cancer cells. Asian Pac J Cancer Prev, 16(6)2425-30. https://doi.org/10.7314/apjcp.2015.16.6.2425
[78] Zhu MX, Zhao TY, Li Y. (2023) Insight into the mechanism of DNA methylation and miRNA-mRNA regulatory network in ischemic stroke. Math Biosci Eng, 20(6)10264-83. https://doi.org/10.3934/mbe.2023450
Type
Published
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Issue
Section
License
Copyright (c) 2025 Life Conflux

This work is licensed under a Creative Commons Attribution 4.0 International License.







