Sphingomyelin mediates the association between natural killer cell receptor 2B4 (CD244) and head and neck cancer
DOI:
https://doi.org/10.71321/cdx1k098Keywords:
NK cell, CD244, Head and Neck Cancer, Sphingomyelin, Mendelian RandomizationAbstract
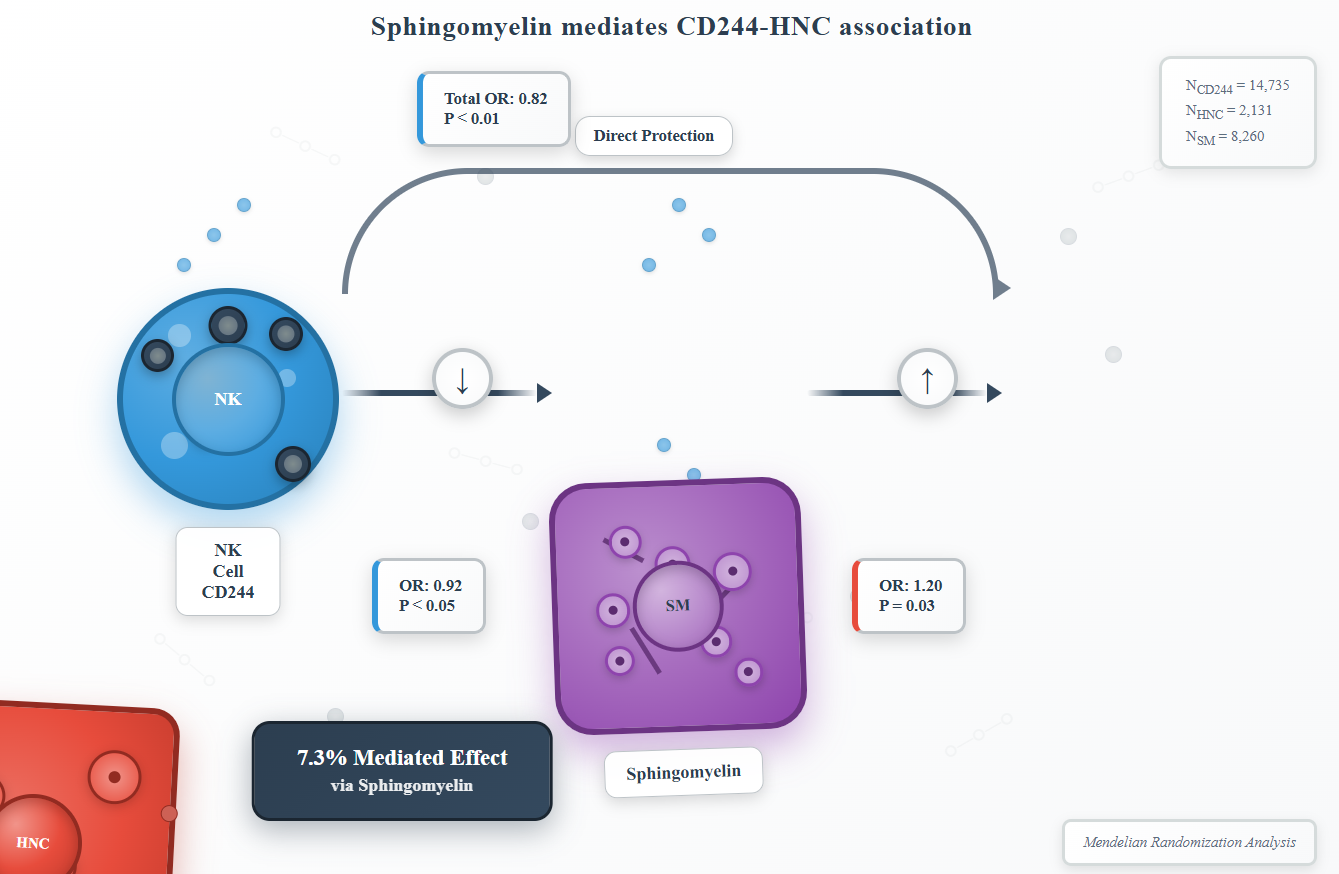
Objective: To investigate the causal link between natural killer (NK) cell receptor CD244 and head and neck cancer (HNC), and to evaluate sphingomyelin (SM) as a potential mediator of this relationship.
Methods: A two-sample Mendelian randomization (MR) analysis was conducted using genome-wide association study (GWAS) summary statistics from European cohorts (NCD244=14,735; NHNC=2,131; NSM=8,260). Instrumental variables (32 SNPs for CD244, 79 SNPs for HNC) were selected at genome-wide significance (P<5×10⁻⁸/5×10⁻⁶) after rigorous linkage disequilibrium clumping. Causal estimates were generated using inverse variance weighting (IVW), MR-Egger, and weighted median methods. Mediation analysis quantified SM's contribution to CD244-HNC associations.
Results: Genetically predicted CD244 levels showed a protective effect against HNC (IVW OR=0.82 per SD, 95% CI:0.71-0.94, P<0.01), while elevated CD244 was associated with reduced SM levels (IVW OR=0.92, 95% CI:0.86-0.98, P<0.05). Conversely, SM demonstrated risk-enhancing effects on HNC (IVW OR=1.20, 95% CI:1.01-1.41, P=0.03). Mediation analysis revealed that SM accounted for 7.3% of the total CD244-associated HNC risk.
Conclusion: This study establishes a causal protective role of CD244 in HNC pathogenesis, partially mediated through SM. The findings highlight CD244 and SM metabolism as potential targets for HNC immunomodulatory therapies. Future research should validate these mechanisms in diverse populations and explore additional mediators.
References
[1] Nersesian S, Carter EB, Lee SN, Westhaver LP, Boudreau JE. Killer instincts: natural killer cells as multifactorial cancer immunotherapy. Front Immunol. 2023 Nov 28;14:1269614. https://doi.org/10.3389/fimmu.2023.1269614.
[2] Mathew SO, Rao KK, Kim JR, Bambard ND, Mathew PA. Functional role of human NK cell receptor 2B4 (CD244) isoforms. Eur J Immunol. 2009 Jun;39(6):1632-41. https://doi.org/10.1002/eji.200838733.
[3] Veillette A, Latour S. The SLAM family of immune-cell receptors. Curr Opin Immunol. 2003 Jun;15(3):277-85. https://doi.org/10.1016/s0952-7915(03)00041-4.
[4] Brown MH, Boles K, van der Merwe PA, Kumar V, Mathew PA, Barclay AN. 2B4, the natural killer and T cell immunoglobulin superfamily surface protein, is a ligand for CD48. J Exp Med. 1998 Dec 7;188(11):2083-90. https://doi.org/10.1084/jem.188.11.2083.
[5] Latchman Y, McKay PF, Reiser H. Identification of the 2B4 molecule as a counter-receptor for CD48. J Immunol. 1998 Dec 1;161(11):5809-12. https://doi.org/10.4049/jimmunol.161.11.5809.
[6] Chuang SS, Kim MH, Johnson LA, Albertsson P, Kitson RP, Nannmark U, et al. 2B4 stimulation of YT cells induces natural killer cell cytolytic function and invasiveness. Immunology. 2000 Jul;100(3):378-83. https://doi.org/10.1046/j.1365-2567.2000.00031.x.
[7] Mathew SO, Kumaresan PR, Lee JK, Huynh VT, Mathew PA. Mutational analysis of the human 2B4 (CD244)/CD48 interaction: Lys68 and Glu70 in the V domain of 2B4 are critical for CD48 binding and functional activation of NK cells. J Immunol. 2005 Jul 15;175(2):1005-13. https://doi.org/10.4049/jimmunol.175.2.1005.
[8] Wu Y, Kuang DM, Pan WD, Wan YL, Lao XM, Wang D, et al. Monocyte/macrophage-elicited natural killer cell dysfunction in hepatocellular carcinoma is mediated by CD48/2B4 interactions. Hepatology. 2013 Mar;57(3):1107-16. https://doi.org/10.1002/hep.26192.
[9] Pende D, Meazza R, Marcenaro S, Aricò M, Bottino C. 2B4 dysfunction in XLP1 NK cells: More than inability to control EBV infection. Clin Immunol. 2019 Jul;204:31-36. https://doi.org/10.1016/j.clim.2018.10.022.
[10] Schlaphoff V, Lunemann S, Suneetha PV, Jaroszewicz J, Grabowski J, Dietz J, et al. Dual function of the NK cell receptor 2B4 (CD244) in the regulation of HCV-specific CD8+ T cells. PLoS Pathog. 2011 May;7(5):e1002045. https://doi.org/10.1371/journal.ppat.1002045.
[11] Chow LQM. Head and Neck Cancer. N Engl J Med. 2020 Jan 2;382(1):60-72. https://doi.org/10.1056/NEJMra1715715.
[12] Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021 May;71(3):209-249. https://doi.org/10.3322/caac.21660.
[13] Rasheduzzaman M, Kulasinghe A, Dolcetti R, Kenny L, Johnson NW, Kolarich D, et al. Protein glycosylation in head and neck cancers: From diagnosis to treatment. Biochim Biophys Acta Rev Cancer. 2020 Dec;1874(2):188422. https://doi.org/10.1016/j.bbcan.2020.188422.
[14] Mehanna H, Paleri V, West CM, Nutting C. Head and neck cancer--Part 1: Epidemiology, presentation, and prevention. BMJ. 2010 Sep 20;341:c4684. https://doi.org/10.1136/bmj.c4684.
[15] Slotte JP. Biological functions of sphingomyelins. Prog Lipid Res. 2013 Oct;52(4):424-37. https://doi.org/10.1016/j.plipres.2013.05.001.
[16] Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization. JAMA. 2017 Nov 21;318(19):1925-1926. https://doi.org/10.1001/jama.2017.17219.
[17] Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014 Sep 15;23(R1):R89-98. https://doi.org/10.1093/hmg/ddu328.
[18] Zhao JH, Stacey D, Eriksson N, Macdonald-Dunlop E, Hedman ÅK, Kalnapenkis A, et al. Genetics of circulating inflammatory proteins identifies drivers of immune-mediated disease risk and therapeutic targets. Nat Immunol. 2023 Sep;24(9):1540-1551. https://doi.org/10.1038/s41590-023-01588-w.
[19] Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023 Jan;613(7944):508-518. doi:10.1038/s41586-022-05473-8.
[20] Chen Y, Lu T, Pettersson-Kymmer U, Stewart ID, Butler-Laporte G, Nakanishi T, et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat Genet. 2023 Jan;55(1):44-53. https://doi.org/10.1038/s41588-022-01270-1.
[21] 1000 Genomes Project Consortium, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010 Oct 28;467(7319):1061-73. https://doi.org/10.1038/nature09534.
[22] Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018 May 30;7:e34408. https://doi.org/10.7554/eLife.34408.
[23] Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res. 2017 Oct;26(5):2333-2355. https://doi.org/10.1177/0962280215597579.
[24] Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016 May;40(4):304-14. https://doi.org/10.1002/gepi.21965.
[25] Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017 Dec 1;46(6):1985-1998. https://doi.org/10.1093/ije/dyx102.
[26] Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017 Dec 1;46(6):1734-1739. https://doi.org/10.1093/ije/dyx034.
[27] Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017 May;32(5):377-389. https://doi.org/10.1007/s10654-017-0255-x.
[28] Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018 May;50(5):693-698. https://doi.org/10.1038/s41588-018-0099-7.
[29] Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013 Nov;37(7):658-65. https://doi.org/10.1002/gepi.21758.
[30] Zhang Y, Liu Z, Choudhury T, Cornelis MC, Liu W. Habitual coffee intake and risk for nonalcoholic fatty liver disease: a two-sample Mendelian randomization study. Eur J Nutr. 2021 Jun;60(4):1761-1767. https://doi.org/10.1007/s00394-020-02369-z.
[31] Carter AR, Sanderson E, Hammerton G, Richmond RC, Davey Smith G, Heron J, et al. Mendelian randomisation for mediation analysis: current methods and challenges for implementation. Eur J Epidemiol. 2021 May;36(5):465-478. https://doi.org/10.1007/s10654-021-00757-1.
[32] Gao P, Lu W, Hu S, Zhao K. Differentially Infiltrated Identification of Novel Diagnostic Biomarkers Associated with Immune Infiltration in Nasopharyngeal Carcinoma. Dis Markers. 2022 Nov 17;2022:3934704. https://doi.org/10.1155/2022/3934704.
[33] Tan L, Cheng D, Wen J, Huang K, Zhang Q. Identification of prognostic hypoxia-related genes signature on the tumor microenvironment in esophageal cancer. Math Biosci Eng. 2021 Sep 7;18(6):7743-7758. https://doi.org/10.3934/mbe.2021384.
[34] Chiha J, Mitchell P, Gopinath B, Burlutsky G, Plant A, Kovoor P, et al. Prediction of Coronary Artery Disease Extent and Severity Using Pulse Wave Velocity. PLoS One. 2016 Dec 22;11(12):e0168598. https://doi.org/10.1371/journal.pone.0168598.
[35] Altvater B, Landmeier S, Pscherer S, Temme J, Schweer K, Kailayangiri S, et al. 2B4 (CD244) signaling by recombinant antigen-specific chimeric receptors costimulates natural killer cell activation to leukemia and neuroblastoma cells. Clin Cancer Res. 2009 Aug 1;15(15):4857-66. https://doi.org/10.1158/1078-0432.CCR-08-2810.
[36] Agresta L, Lehn M, Lampe K, Cantrell R, Hennies C, Szabo S, et al. CD244 represents a new therapeutic target in head and neck squamous cell carcinoma. J Immunother Cancer. 2020 Mar;8(1):e000245. https://doi.org/10.1136/jitc-2019-000245.
[37] Wang H, Luo Y, Chen H, Hou H, Hu Q, Ji M. Non-Targeted Serum Lipidomics Analysis and Potential Biomarkers of Laryngeal Cancer Based on UHPLC-QTOF-MS. Metabolites. 2022 Nov 9;12(11):1087. https://doi.org/10.3390/metabo12111087.
[38] Zheng X, Hou Z, Qian Y, Zhang Y, Cui Q, Wang X, et al. Tumors evade immune cytotoxicity by altering the surface topology of NK cells. Nat Immunol. 2023 May;24(5):802-813. https://doi.org/10.1038/s41590-023-01462-9.
[39] Farhangnia P, Ghomi SM, Mollazadehghomi S, Nickho H, Akbarpour M, Delbandi AA. SLAM-family receptors come of age as a potential molecular target in cancer immunotherapy. Front Immunol. 2023 May 11;14:1174138. https://doi.org/10.3389/fimmu.2023.1174138.
[40] Jacobi J, García-Barros M, Rao S, Rotolo JA, Thompson C, Mizrachi A, et al. Targeting acid sphingomyelinase with anti-angiogenic chemotherapy. Cell Signal. 2017 Jan;29:52-61. https://doi.org/10.1016/j.cellsig.2016.09.010.
Type
Published
Data Availability Statement
The genetic data of circulating inflammatory proteins and plasma metabolites used in this article are derived from two studies published in Nature (doi:10.1038/s41590-023-01588-w and 10.1038/s41588-022-01270-1). The genetic data of HNC (Head and Neck Cancer) patients are sourced from the Finngen database, with detailed descriptions available in the Materials section.
Issue
Section
License
Copyright (c) 2025 Life Conflux

This work is licensed under a Creative Commons Attribution 4.0 International License.







