Mendelian Randomization Reveals Height as a Risk Factor and Potential Therapeutic Target for Testicular Cancer
DOI:
https://doi.org/10.71321/sqcqnh24Keywords:
Height, Testicular cancer, Mendelian randomization, Bayesian colocalization, Susceptibility geneAbstract
Objective
To investigate the potential causal relationship between height and the risk of developing testicular cancer using Mendelian Randomization analysis.
Methods
We utilized phenotype data on height and testicular cancer from two European ancestry cohorts, integrating data from the IEU, FinnGen, and UK Biobank databases. Linkage Disequilibrium Score regression and SNP-associated gene enrichment analyses were initially conducted to assess the association between SNPs and the phenotypes. A two-sample MR approach was then applied to evaluate the causal relationship between height and testicular cancer risk. An additional cohort was analyzed for validation, followed by a meta-analysis to combine results. Reverse MR and colocalization analyses were performed to investigate potential reverse causality. Gene enrichment analysis was conducted to elucidate the biological mechanisms linking height to testicular cancer, and potential eQTL targets for testicular cancer were explored using summary-data-based MR and colocalization analysis.
Results
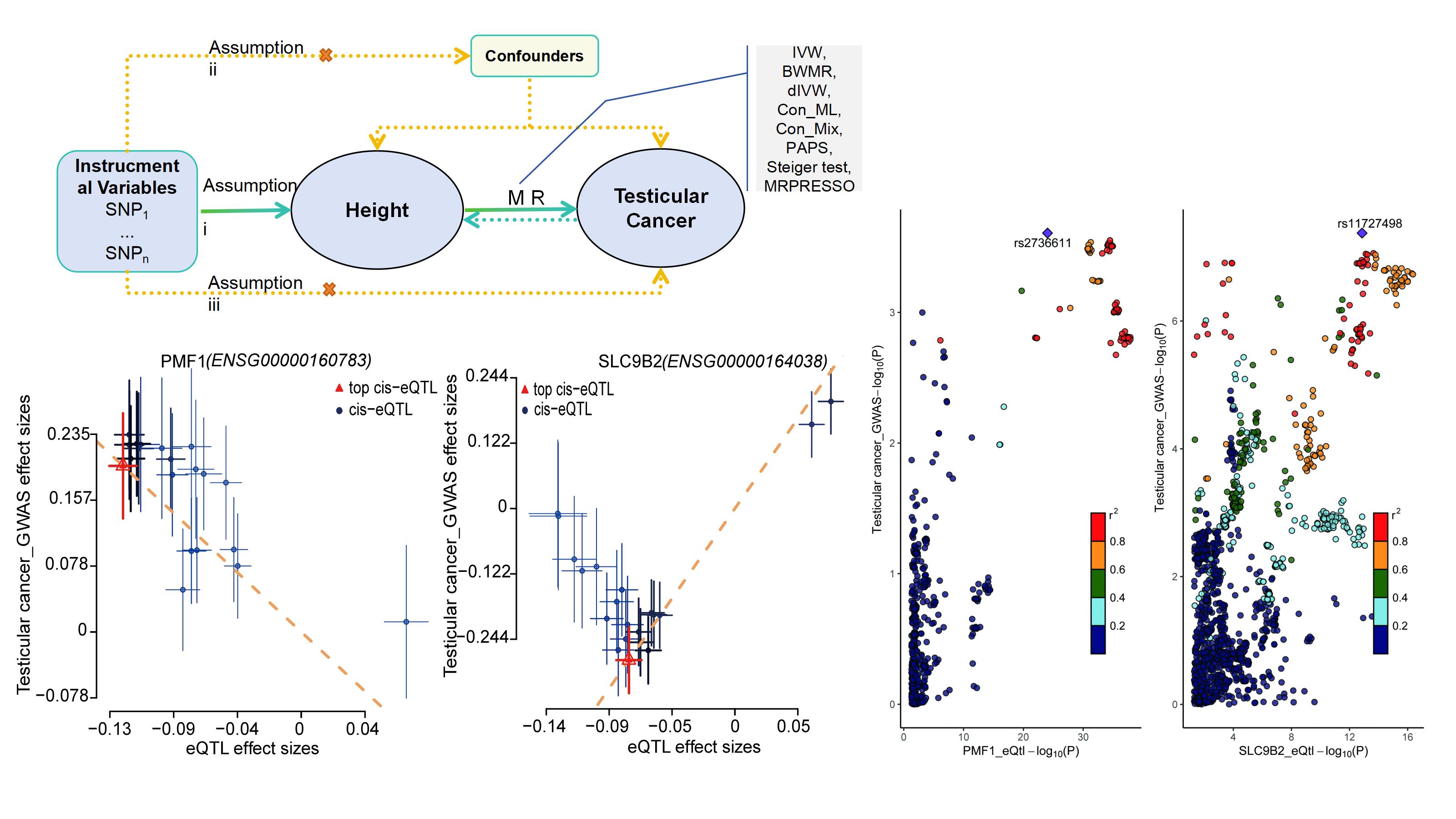
The Inverse Variance Weighted method revealed a significant association between genetically predicted height and testicular cancer risk, with an odds ratio of 1.372 (95% CI: 1.024-1.837, p-value < 0.05). Gene enrichment analysis suggested that the extracellular matrix-related pathway might underlie the increased risk of testicular cancer associated with height. PMF1 and SLC9B2 were identified as potential targets for testicular cancer from summary-data-based MR, colocalization, and gene expression analysis.
Conclusions
This Mendelian randomization study provides evidence supporting a causal relationship between height and the risk of developing testicular cancer, with PMF1 and SLC9B2 identified as potential targets for further investigation.
References
[1] Yu S, Guo Z, Qiu Z, Wang L, Chen X, & Xuan F. (2024). Global burden and trends of testicular cancer in adolescents and young adults from 1990 to 2021, with predictions to 2035. Scientific Reports, 14(1), 31787. https://doi.org/10.1038/s41598-024-82897-4
[2] Chavarriaga J, Nappi L, Papachristofilou A, Conduit C, & Hamilton RJ. (2025). Testicular cancer. Lancet, 406(10498), 76-90. https://doi.org/10.1016/s0140-6736(25)00455-6
[3] Bhanushali C, Shah RN, Vojjala N, Jayakumar J, Harisingani AR, & Jani C. (2025). Racial and socioeconomic disparities in testicular cancer survival outcomes: A SEER database analysis. Journal of Clinical Oncology, 43(16_suppl), 5028-5028. https://doi.org/10.1200/JCO.2025.43.16_suppl.5028
[4] Tateo V, Thompson ZJ, Gilbert SM, Cortessis VK, Daneshmand S, Masterson TA, et al. (2025). Epidemiology and Risk Factors for Testicular Cancer: A Systematic Review. Eur Urol, 87(4), 427-441. https://doi.org/10.1016/j.eururo.2024.10.023
[5] Yazici S, Del Biondo D, Napodano G, Grillo M, Calace FP, Prezioso D, et al. (2023). Risk Factors for Testicular Cancer: Environment, Genes and Infections-Is It All? Medicina (Kaunas), 59(4). https://doi.org/10.3390/medicina59040724
[6] Levy M, Hall D, Sud A, Law P, Litchfield K, Dudakia D, et al. (2017). Mendelian randomisation analysis provides no evidence for a relationship between adult height and testicular cancer risk. Andrology, 5(5), 914-922. https://doi.org/10.1111/andr.12388
[7] Yarmolinsky J, Wade KH, Richmond RC, Langdon RJ, Bull CJ, Tilling KM, et al. (2018). Causal Inference in Cancer Epidemiology: What Is the Role of Mendelian Randomization? Cancer Epidemiol Biomarkers Prev, 27(9), 995-1010. https://doi.org/10.1158/1055-9965.Epi-17-1177
[8] Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, et al. (2022). Mendelian randomization. Nature Reviews Methods Primers, 2(1), 6. https://doi.org/10.1038/s43586-021-00092-5
[9] Burgess S, Small DS, & Thompson SG. (2017). A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res, 26(5), 2333-2355. https://doi.org/10.1177/0962280215597579
[10] Smith GD, & Ebrahim S. (2003). 'Mendelian randomization': can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol, 32(1), 1-22. https://doi.org/10.1093/ije/dyg070
[11] Lawlor DA, Harbord RM, Sterne JA, Timpson N, & Davey Smith G. (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med, 27(8), 1133-1163. https://doi.org/10.1002/sim.3034
[12] Davey Smith G, & Hemani G. (2014). Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet, 23(R1), R89-98. https://doi.org/10.1093/hmg/ddu328
[13] Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. (2021). Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. Jama, 326(16), 1614-1621. https://doi.org/10.1001/jama.2021.18236
[14] Võsa U, Claringbould A, Westra HJ, Bonder MJ, Deelen P, Zeng B, et al. (2021). Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet, 53(9), 1300-1310. https://doi.org/10.1038/s41588-021-00913-z
[15] Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature, 491(7422), 56-65. https://doi.org/10.1038/nature11632
[16] Zheng J, Erzurumluoglu AM, Elsworth BL, Kemp JP, Howe L, Haycock PC, et al. (2017). LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics, 33(2), 272-279. https://doi.org/10.1093/bioinformatics/btw613
[17] Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, et al. (2015). An atlas of genetic correlations across human diseases and traits. Nat Genet, 47(11), 1236-1241. https://doi.org/10.1038/ng.3406
[18] Li L, Fu L, Zhang L, & Feng Y. (2022). Mendelian randomization study of the genetic interaction between psoriasis and celiac disease. Sci Rep, 12(1), 21508. https://doi.org/10.1038/s41598-022-25217-y
[19] Burgess S, & Thompson SG. (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol, 40(3), 755-764. https://doi.org/10.1093/ije/dyr036
[20] Levin MG, Judy R, Gill D, Vujkovic M, Verma SS, Bradford Y, et al. (2020). Genetics of height and risk of atrial fibrillation: A Mendelian randomization study. PLoS Med, 17(10), e1003288. https://doi.org/10.1371/journal.pmed.1003288
[21] Hemani G, Tilling K, & Davey Smith G. (2017). Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet, 13(11), e1007081. https://doi.org/10.1371/journal.pgen.1007081
[22] Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. (2016). Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet, 48(5), 481-487. https://doi.org/10.1038/ng.3538
[23] Burgess S, Scott RA, Timpson NJ, Davey Smith G, & Thompson SG. (2015). Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol, 30(7), 543-552. https://doi.org/10.1007/s10654-015-0011-z
[24] Verbanck M, Chen CY, Neale B, & Do R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet, 50(5), 693-698. https://doi.org/10.1038/s41588-018-0099-7
[25] Zhao J, Ming J, Hu X, Chen G, Liu J, & Yang C. (2020). Bayesian weighted Mendelian randomization for causal inference based on summary statistics. Bioinformatics, 36(5), 1501-1508. https://doi.org/10.1093/bioinformatics/btz749
[26] Burgess S, Foley CN, Allara E, Staley JR, & Howson JMM. (2020). A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat Commun, 11(1), 376. https://doi.org/10.1038/s41467-019-14156-4
[27] Yu K, Chen XF, Guo J, Wang S, Huang XT, Guo Y, et al. (2023). Assessment of bidirectional relationships between brain imaging-derived phenotypes and stroke: a Mendelian randomization study. BMC Med, 21(1), 271. https://doi.org/10.1186/s12916-023-02982-9
[28] Yin Q, & Zhu L. (2024). Does co-localization analysis reinforce the results of Mendelian randomization? Brain, 147(1), e7-e8. https://doi.org/10.1093/brain/awad295
[29] Ting Y, Jun S, & Hyunseung K. (2021). Debiased inverse-variance weighted estimator in two-sample summary-data Mendelian randomization. The Annals of Statistics, 49(4), 2079-2100. https://doi.org/10.1214/20-AOS2027
[30] Mounier N, & Kutalik Z. (2023). Bias correction for inverse variance weighting Mendelian randomization. Genet Epidemiol, 47(4), 314-331. https://doi.org/10.1002/gepi.22522
[31] Burgess S, Butterworth A, & Thompson SG. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol, 37(7), 658-665. https://doi.org/10.1002/gepi.21758
[32] Bowden J, Davey Smith G, & Burgess S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol, 44(2), 512-525. https://doi.org/10.1093/ije/dyv080
[33] Wu Y, Zeng J, Zhang F, Zhu Z, Qi T, Zheng Z, et al. (2018). Integrative analysis of omics summary data reveals putative mechanisms underlying complex traits. Nat Commun, 9(1), 918. https://doi.org/10.1038/s41467-018-03371-0
[34] Rafael Newlands F, Natalia Bonfim dS, Marcio Sidney DBF, Fernanda Ferreira L, Arovel Oliveira MJ, Rafael Texeira B, et al. (2022). Screening pediatric testicular cancer: A literature review. Archives of Community Medicine and Public Health.
[35] Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol, 4(2), 293-305. https://doi.org/10.1038/s41564-018-0306-4
[36] Battle A, Brown CD, Engelhardt BE, & Montgomery SB. (2017). Genetic effects on gene expression across human tissues. Nature, 550(7675), 204-213. https://doi.org/10.1038/nature24277
[37] Rasooly D, Peloso GM, Pereira AC, Dashti H, Giambartolomei C, Wheeler E, et al. (2023). Genome-wide association analysis and Mendelian randomization proteomics identify drug targets for heart failure. Nat Commun, 14(1), 3826. https://doi.org/10.1038/s41467-023-39253-3
[38] Ding R, Zou X, Qin Y, Gong L, Chen H, Ma X, et al. (2023). xQTLbiolinks: a comprehensive and scalable tool for integrative analysis of molecular QTLs. Brief Bioinform, 25(1), bbad440. https://doi.org/10.1093/bib/bbad440
[39] Yu G, Wang LG, Han Y, & He QY. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics, 16(5), 284-287. https://doi.org/10.1089/omi.2011.0118
[40] Kolberg L, Raudvere U, Kuzmin I, Adler P, Vilo J, & Peterson H. (2023). g:Profiler-interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res, 51(W1), W207-w212. https://doi.org/10.1093/nar/gkad347
[41] Pierce BL, Ahsan H, & Vanderweele TJ. (2011). Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol, 40(3), 740-752. https://doi.org/10.1093/ije/dyq151
[42] Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, et al. (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet, 47(3), 291-295. https://doi.org/10.1038/ng.3211
[43] Lerro CC, McGlynn KA, & Cook MB. (2010). A systematic review and meta-analysis of the relationship between body size and testicular cancer. Br J Cancer, 103(9), 1467-1474. https://doi.org/10.1038/sj.bjc.6605934
[44] Choi YJ, Lee DH, Han KD, Yoon H, Shin CM, Park YS, et al. (2019). Adult height in relation to risk of cancer in a cohort of 22,809,722 Korean adults. Br J Cancer, 120(6), 668-674. https://doi.org/10.1038/s41416-018-0371-8
[45] Nunney L. (2013). The real war on cancer: the evolutionary dynamics of cancer suppression. Evol Appl, 6(1), 11-19. https://doi.org/10.1111/eva.12018
[46] Rowlands MA, Gunnell D, Harris R, Vatten LJ, Holly JM, & Martin RM. (2009). Circulating insulin-like growth factor peptides and prostate cancer risk: a systematic review and meta-analysis. Int J Cancer, 124(10), 2416-2429. https://doi.org/10.1002/ijc.24202
[47] Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. (2010). Common SNPs explain a large proportion of the heritability for human height. Nat Genet, 42(7), 565-569. https://doi.org/10.1038/ng.608
[48] Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, Zusmanovich P, et al. (2008). Many sequence variants affecting diversity of adult human height. Nat Genet, 40(5), 609-615. https://doi.org/10.1038/ng.122
[49] Qian F, & Huo D. (2020). Circulating Insulin-Like Growth Factor-1 and Risk of Total and 19 Site-Specific Cancers: Cohort Study Analyses from the UK Biobank. Cancer Epidemiol Biomarkers Prev, 29(11), 2332-2342. https://doi.org/10.1158/1055-9965.Epi-20-0743
[50] Fanti M, & Longo VD. (2024). Nutrition, GH/IGF-1 signaling, and cancer. Endocr Relat Cancer, 31(11). https://doi.org/10.1530/erc-23-0048
[51] Pitetti JL, Calvel P, Zimmermann C, Conne B, Papaioannou MD, Aubry F, et al. (2013). An essential role for insulin and IGF1 receptors in regulating sertoli cell proliferation, testis size, and FSH action in mice. Mol Endocrinol, 27(5), 814-827. https://doi.org/10.1210/me.2012-1258
[52] Li L, Huang J, & Liu Y. (2023). The extracellular matrix glycoprotein fibrillin-1 in health and disease. Front Cell Dev Biol, 11, 1302285. https://doi.org/10.3389/fcell.2023.1302285
[53] Wang W, Rigueur D, & Lyons KM. (2014). TGFβ signaling in cartilage development and maintenance. Birth Defects Res C Embryo Today, 102(1), 37-51. https://doi.org/10.1002/bdrc.21058
[54] Stephenson A, Bass EB, Bixler BR, Daneshmand S, Kirkby E, Marianes A, et al. (2024). Diagnosis and Treatment of Early-Stage Testicular Cancer: AUA Guideline Amendment 2023. J Urol, 211(1), 20-25. https://doi.org/10.1097/ju.0000000000003694
[55] New SNPs from Testicular Cancer GWAS. (2017). Cancer Discov, 7(9), Of5. https://doi.org/10.1158/2159-8290.Cd-nb2017-103
[56] Pluta J, Pyle LC, Nead KT, Wilf R, Li M, Mitra N, et al. (2021). Identification of 22 susceptibility loci associated with testicular germ cell tumors. Nat Commun, 12(1), 4487. https://doi.org/10.1038/s41467-021-24334-y
[57] Zhou X, Jiang M, Liu Z, Xu M, Chen N, Wu Z, et al. (2021). Na(+)/H(+)-Exchanger Family as Novel Prognostic Biomarkers in Colorectal Cancer. J Oncol, 2021, 3241351. https://doi.org/10.1155/2021/3241351
[58] Lu X, Luo Y, Nie X, Zhang B, Wang X, Li R, et al. (2023). Single-cell multi-omics analysis of human testicular germ cell tumor reveals its molecular features and microenvironment. Nature Communications, 14(1), 8462. https://doi.org/10.1038/s41467-023-44305-9
[59] Battaglino RA, Pham L, Morse LR, Vokes M, Sharma A, Odgren PR, et al. (2008). NHA-oc/NHA2: a mitochondrial cation-proton antiporter selectively expressed in osteoclasts. Bone, 42(1), 180-192. https://doi.org/10.1016/j.bone.2007.09.046
Type
Published
Data Availability Statement
The datasets analyzed during the current study are available in the IEU Open GWAS repository (https://gwas.mrcieu.ac.uk/). Data on TC in cohort 1 can be found in the FinnGen project (https://www.finngen.fi/).
Issue
Section
License
Copyright (c) 2025 Life Conflux

This work is licensed under a Creative Commons Attribution 4.0 International License.







